Automatic Human Sperm Concentration in Microscopic Videos
Type of article: Original
1 Email: karima.boumaza@univ-usto.dz
Abstract
Background: The process of counting human sperm cell studies are of noteworthy
interest to biologists researching sperm function and to medical practitioners
in charge of mitigating male infertility. Currently, this assessment is
performed manually by observing the sperm samples through a phase-contrast
microscope using expert knowledge to do a subjective quality judgment.
Aims: To
eliminate the subjective and error-prone influences of the manual semen
exploration and to evade inter and intra-laboratory discrepancies in semen
analysis test results
Methods:
This paper, introduces a Computer-Assisted Sperm Analysis (CASA) to infer the
concentration of human sperm in three steps: (i) the human sperm pretreatment
to be investigated by videos acquired using a microscopic, which consists of a
conversion the RGB into the YCbCr color space, the Gaussian filter along with
the Discrete Wavelet Filtering (DWF); (ii) segmenting the image in twofold
classes: spermatozoa and the background, followed by the Sobel edge detection
detector to produce these outcomes; and (iii) distinguishing true sperm from
false ones with a classification technique consisting of decision trees and
relying on invariant features: the dimensions of the spermatozoid head bounding
ellipse as well as its surface.
Results: To
test the robustness of the recommended system, the outcomes from automatic and
manual tests have been compred compared. The manual tests have been done by
three andrologists. There has been real improvement of precision as well as
treatment time, which make this framework useful for groups who intend to
design new CASA systems.
Conclusion: In this study, a simple system for automatic concentration assessment of
spermatozoa founded on image processing techniques is proposed and implemented.
Keywords:
Decision Trees, Discrete Wavelet Transform, Sobel Filter, Human Sperm,
Computer-Assisted Sperm Analysis (CASA), Sperm Classification.
Corresponding author:Karima Boumaza University of Science and Technology
Oran, Oran, Algeria, email karima.boumaza@univ-usto.dz
Received: 30 June, 2018, Accepted: 01 December, 2018, English editing: 30 December,
2018,Published: 31 December, 2018.
Screened by iThenticate.©2017-2018 KNOWLEDGE
KINGDOM PUBLISHING.
1. INTRODUCTION
Infertility cases have shown
an increasing boost in recent years (1). It can impact unfavorably the quality
of a couple’s life and causes social, as well as emotional problems (2) as
stress, depression and sexual apathy (3). Male infertility results most
commonly from deficiencies in the semen and the conservative criteria for semen
quality. Semen analysis test is required as an initial and most vital stage for
male factor infertility appraisal besides treatment therapy determination. This
test included a physical examination, hormonal evaluation, sperm parameter
determination and genetic analysis (1).The conventional appraisal of sperm
parameters at fertility clinics in addition to research laboratories is
strenuous and subjective (4) with substantial intra- and inter-laboratory
changeability. Typically, the technicians use microscopes to count sperm cells
manually. To replace these subjective assessment methods, Computer-Assisted
Sperm Analysis (CASA) frameworks are from the 1980s. They are usually thought
to deliver objective with repeatable results for semen analysis (5). However
the methods used behind these systems are not openly accessed and the results
of some CASA were not encouraging enough for some samples. Thereafter, many
studies and researches have improved it. An important standard sperm parameter
is the sperm concentration or semen density. It is the oldest reported to be
investigated during a semen analysis (6). It is reported in sperm/milliliter
(mL). According to the WHO (7), the concentration in a normal simple is 15×106
mL and a low concentration is defined as Oligozoospermia.
2. MATERIALS AND METHODS
2.1- Materials:
The experiment dataset is composed of microscopic video sequence representing
sperm motility. To evaluate the proposed CASA system, some sequences have been
picked from the dataset used from (8) that contains video recordings
corresponding to sperm samples of 30 patients accomplished employing a
phase-contrast microscope with an enlargement of ×120 at the Isfahan Fertility
and Infertility Center. These video sequences have resolution as well as frame
rate of 240 × 320 pixels and 25 fps, correspondingly.
2.2- Methods:
The non-uniform
illumination, low contrast, small size of the microscopic images, a high number
of sperms and human visual problem, an automated method for sperm concentration
is required. As a contribution in the fertility area, the proposed CASA system
aims to automate the procedure for measuring the sperm concentration by probing
and analyzing a microscopic video sequence of a sperm sample taken according to
WHO guidelines and standards (7). As shown in Figure 71, the designed CASA
process operates in three steps:
1. The first module performs
the pre-treatment of raw microscopic videos, the color space conversion, the
contrast enhancement, and the noise reduction phase;
2. The second module
separates the spermatozoa from other impurities (debris, seminal fluid,
electronic noise) by applying the Sobel boundary detector that leads to a
binary image containing white blobs of different sizes. An output image
containing the spermatozoa is isolated by means of a thresholding operation on
the blobs.
3. The third module receives
the output result from the second module, the segmented objects are not all
spermatozoa. A classification phase of these objects is necessary. Knowing that
the head of spermatozoid has an elliptical shape and almost identical size, two
features have been chosen as discriminatory and used in the two-class
classification process. The result obtained at this module output is the total
number of spermatozoa which means the concentration.
2.2.1 Noise
reduction:
Like any artificial vision
system, the pre-processing of raw images is a very important step because the
whole system has it precision based on it.
The poor quality of microscopic images of sperm led us to proceed in
several steps. We have been partially inspired by the work in (9,10).
1) Conversion
from RGB color space to YCbCr
We have as input a
microscopic video recorded 240 × 320 pixels with a frequency of 25 fps. First,
an RGB-to-YCbCr color space conversion for each frame of video is required (9).
Because of the similarity between the Luminance (Y) component and the original
grayscale imageries, this component is used for the subsequent system stage.
The expression for performing this conversion is given below (9):
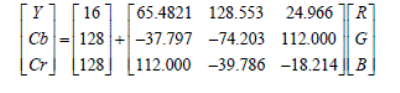
2) Smoothing
with a 2D Gaussian Filter
In order to blur the image
and reduce details and noise caused by random electrical disturbance in video
imaging system, a Gaussian filter was applied on the resulting image. The Gaussian probability distribution function
(pdf) for a 1D random variable with mean m and standard deviation s is
given by

For a mean vector m and a covariance matrix Σ, the pdf
for a multivariate normal is
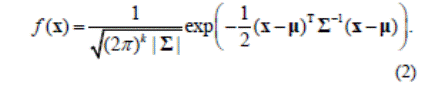
3) Filtering
with the Discrete Wavelet Transform
Finally, the de-noising is
performed using the two-dimensional wavelet transformation. The speed of
calculation, even for relatively high decomposition orders, and the good
discernibility of the structures turns the framework very effective and widely
tool used for reducing digital image noise. This transformation favors a local
and non-global study of the image: decomposition is not done in the periodic
functions’ space but with another class of functions such as Daubechies, Haar,
Coiflets, Symmlets .
After the application of the
aforementioned wavelet transform, the Coiflet 2 can be used and this
decomposition occurs at level 4 because the noise signals affecting the input
images can be extracted at a satisfactory rate while handling appropriate
images for supplementary analysis.
2.2.2. Sperm edge detection:
The process continued with
edge detection step. The image at the entrance of this module will therefore go
through two competing treatments as follows:
-
Median filtering (3 x 3) to suppress impulse
noises.
-
Sobel edge detector (3 x 3), which performs a
2-D spatial gradient measurement on an image and emphasizes regions of high
spatial frequency that correspond to edges (9). So as a result, we get a
contours image.
2.2.3. Features
extraction
The third module output
image is binary and includes blobs having similar sizes to spermatozoa's but
with various shapes. To leave only real spermatozoa, this module has for role
to identify them and erase the other objects. The adopted solution lies in the
exploitation of the elliptical form of the spermatozoid‘s head as well as its
surface. To do this, for each blob, the bounding ellipse features and its
surface are computed (see Figure 1). Bounding ellipses can be found with the
help of the Hough Transform (HT) or investigating the image visually. The
handpicked feature vector is v =[a, b, S]T where, for a given ellipse i, the
discriminating features are the major axis a Î [4.20, 16.90], the minor axis b< 7.74 , and
SÎ [55, 200] is the blob area, all measured in
pixels.
After that, this feature
vector undergoes a basic classification of the type "decision trees"
from Figure 2. At this module completion, the output image undergoes labeling
and counting processes to get the concentration value.

Figure 1. Ellipse Geometry.
As preliminary work of the
classification step, we manually analyzed, with the support of andrology
technicians, several video sequences of our database and manually measured the
limits of the 3 characteristics for spermatozoa. The experimental values found
are:
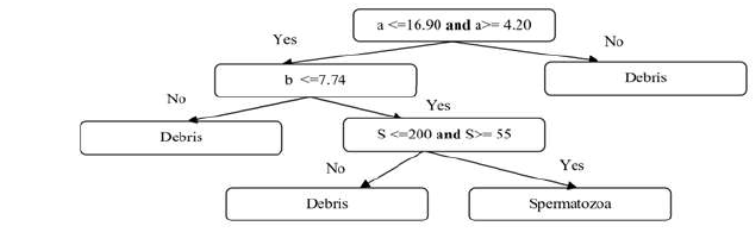
Figure 2. Decision tree
organization
3. RESULTS:
The recommended
classification scheme works with microscopic video, which gives a satisfactory
rate of results in spite of the low quality images (low contrast and small
size) of the microscopic. The systems fragmented the videos into frames, each
frame is processed by our algorithm.
The steps for the noise
reduction stage are exhibited in Figure 3 where Figure 3a has a sperm input
image. After mapping these RGB images into the YCbCr color space, their Y
components become visible as in Figure 3b where noise is explicitly visible.
However, this noise can be alleviated by the Gaussian filter (i.e., reduce the
noise) whose output is in Figure 3c. In this image, the noise is decreased.
Still, this amount of signal contamination is not adequate for further processing.
So applying discrete wavelet transform is required. Figure 3d represent the
output of discrete wavelet transform (DTW).
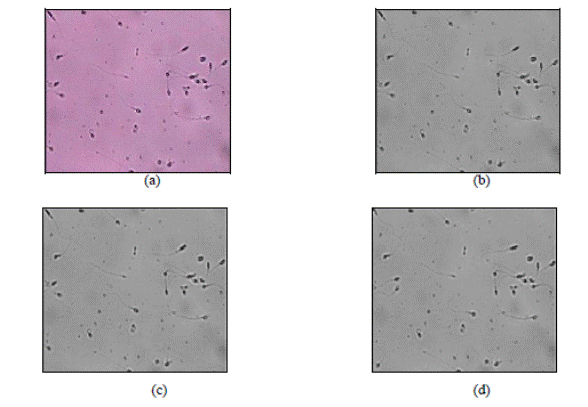
Figure 3. Noise
reduction steps: (a) RGB input image, (b) Image of the Y component (YCbCr
space), (c) Gaussian filtered output, and (d) DWT results.
Figure 4 depicts the edge
detection stage results that come after eradicating the image noise, as in
Figure 4a that contains the output of median filter. The Sobel detector
performs edge detection and its experimental results are in Figure 4b.
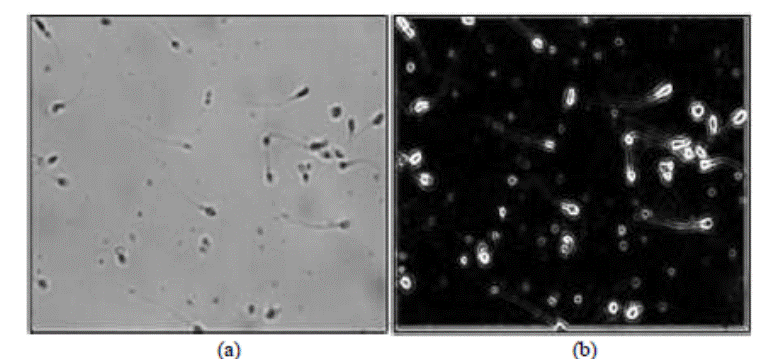
Figure 4. Edge detection step: (a) 3×3 median filtered
image, and (b) Sobel algorithm output.
Figure 5 indicates the elliptical
annotation step for the features extraction. Figure 5a has a complete video
frame encompassing a variety of cells and spermatozoids, so that it
characterizes a sample for algorithm evaluation. In Figure 5b each elliptic
shape corresponds to a detected sperm head.
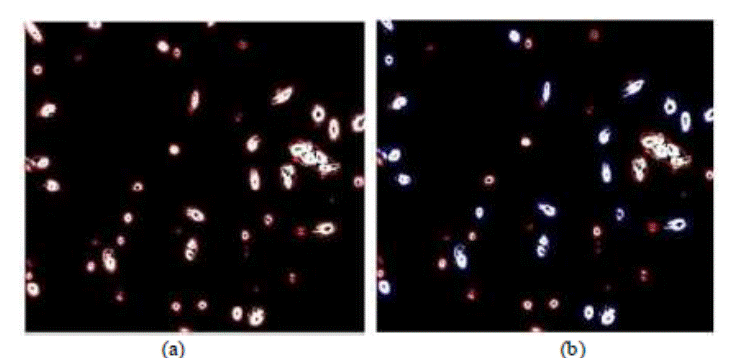
Figure 5. Elliptical annotation and
feature extractions step. (a) The
original image. (b) Detected sperm head with blue
color and debris in red color.
After demonstrating the viability of the recommended framework, the
focus shift to a comparative study with a statistical system evaluation. The
true concentration values are unknown for all videos in the used database.
Moreover, even if a commercial CASA system was available, its results would
also have demanded three experienced andrology experts to measure manually the
values of sperm concentration per video for each one of the 8 video sequences
individually for the available database. The attained results for the manual
analysis by the three experts and those of our system are in Table 1 in
addition to Figure 6.
Table1.Obtained values of concentration by the 3
experts (manual method) and the proposed system (automatic method)
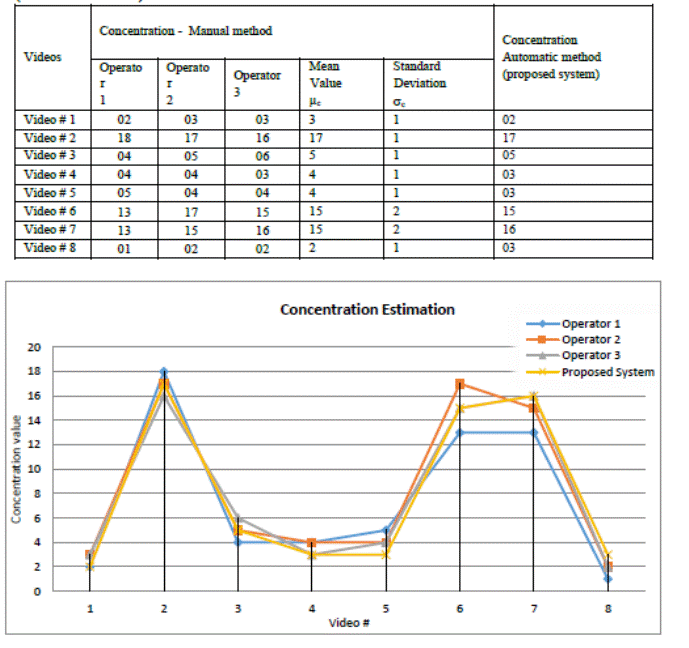
Figure 6. Graphical representation of the obtained values
of concentration by the 3 experts (manual method) and the proposed system
(automatic method).
4. DISCUSSION
In Table 1
received two extra columns concerning the manual analysis results of the 3
operators to define the statistical values of µ and s. We have rounded them because they represent
spermatozoids (whole numbers). After examining Table 1 and Figure 6, the
concentration values obtained with the proposed CASA system (automatic method)
for the 8 video sequences are very precise and close to those obtained manually
and are all in the interval: μc ± sc of the manually measured value. This
statement can be confirmed graphically by looking at the curve representing the automatic
concentration measurements is enveloped by the 3 manual measurements curves to
± sc that appears in Figure 6.
Concluding,
the suggested system gives very good results compared to those obtained
manually with very good algorithm
execution time (nearly 4 times inferior to the time to get a manual analysis by
a human expert) as follows:
- The execution time of our algorithm = 80 seconds; and
- The average time of manipulation analysis by a human expert
= 300 seconds.
5. CONCLUSION
A semen analysis is a vital examination for spotting male infertility. It
measures the concentration, morphology and motility of sperms under the
microscope. While using the manual tests is a laborious and subjective task,
several other works obtained and treated sperm images to obtain results more
objectively. This examination was initially concerned with the automatic
detection of the spermatozoids and the associated counting by implementing a
CASA system relying on microscopic videos of human semen. It makes evident that
the image processing methods, using a decision tree algorithm give a decent classification,
and the accomplished outcomes are clearer, more transparent besides easier to
comprehend with regard to the manual methods. This designed framework is a
substantial achievement towards more sophisticated CASA systems.
Soft computing can be used to reduce problems and augment the number of
CASA functionalities as it is done with other computer-assisted medical
diagnosis (11, 12, 13, 14, 15, 16, 17, 18, 19). Debris and other fine detail
can be investigated with the help of Super-Resolution (15. 19).
ACKNOWLEDGMENT
The authors would like to
thank Mr. Mohammad Reza AHMADZADEH, PhD,
Isfahan University of Technology (Iran) for allowing us to use his database of microscopic sperm videos and his manual
assessment of the used videos.
We would like to thank Dr N.
BENAMAR and Dr. A. BOUALEM, Andrologists at EHU Hospital of Oran (Algeria) for
helping us to do the manual assessment of sperm motility from the videos and their
valuable remarks and suggestions
REFERENCES
(1) HASIKIN, Khairunnisa, ISA, Nor Ashidi Mat, MOHAMED, Mahaneem, et al. A New Region-Based Adaptive Thresholding For Sperm Motility Segmentation. Malaysian Journal of Computer Science, 2017, vol. 29, no 4.
(2) SHAKER, Fariba, MONADJEMI, S. Amirhassan, et NAGHSH-NILCHI, Ahmad Reza. Automatic detection and segmentation of sperm head, acrosome and nucleus in microscopic images of human semen smears. Computer methods and programs in biomedicine, 2016, vol. 132, p. 11-20.
(3) DOMAR, Alice D., BROOME, Alexis, ZUTTERMEISTER, Patricia C., et al. The prevalence and predictability of depression in infertile women. Fertility and sterility, 1992, vol. 58, no 6, p. 1158-1163.
(4) URBANO, Leonardo F., MASSON, Puneet, VERMILYEA, Matthew, et al.Automatic Tracking and Motility Analysis of Human Sperm in Time-Lapse Images. IEEE transactions on medical imaging, 2017, vol. 36, no 3, p. 792-801.
(5) BORYSHPOLETS, S., KOWALSKI, R. K., DIETRICH, G. J., et al. Different computer-assisted sperm analysis (CASA) systems highly influence sperm motility parameters. Theriogenology, 2013, vol. 80, no 7, p. 758-765.
(6)
MORTIMER, David. Practical laboratory
andrology. Oxford University Press on Demand, 1994.
(7) COOPER, Trevor G., NOONAN, Elizabeth, VON ECKARDSTEIN, Sigrid, et al.World Health Organization reference values for human semen characteristics. Human reproduction update, 2010, vol. 16, no 3, p. 231-245.
(8) IMANI, Yoones, TEYFOURI, Niloufar, AHMADZADEH, Mohammad Reza, et al.A new method for multiple sperm cells tracking. Journal of medical signals and sensors, 2014, vol. 4, no 1, p. 35.
(9)
GHASEMIAN, Fatemeh, MIRROSHANDEL, Seyed
Abolghasem, MONJI-AZAD, Sara, et al. An efficient method for automatic
morphological abnormality detection from human sperm images. Journal Computer
methods and programs in biomedicine, 2015, vol. 122, no 3, p. 409-420.
(10)
BOUMAZA, Karima, LOUKIL, Abdelhamid, et
AARIZOU, Kaouthar. Computer Aided Human Sperm Motility Detection.
IEEE
International Conference on Automatic control, Telecommunication and Signals, 2017.
(11) Mahmoodi, M. S., and S. A.
Mahmoodi. “Design of CAD System of Solitary Pulmonary Nodule With Harmony Classification and Fuzzy System”. Medical
Technologies Journal, Vol. 1, no. 4, Nov. 2017, pp. 102-,
doi:https://doi.org/10.26415/2572-004X-vol1iss4p102-102.
(12) Belgherbi, A., I. Hadjidj,
and A. Bessaid. “Computer-Aided Detection of Simultaneous Abdominal Organ from
CT Images Based on Iterative Watershed Transform”. Medical Technologies
Journal, Vol. 1, no. 1, Mar. 2017, pp. 8-8,
doi:https://doi.org/10.26415/2572-004X-vol1iss1p8-8.
(13) Herrmann, A. E. & Estrela,
V. V. (2016). Content-based
image retrieval (CBIR) in remote clinical diagnosis and healthcare. In M.
Cruz-Cunha, I. Miranda, R. Martinho, & R. Rijo (Eds.), Encyclopedia of
E-Health and Telemedicine (pp. 495-520). Hershey, PA: IGI Global.
doi:10.4018/978-1-4666-9978-6.ch039
(14) Jesus, M.A., &
Estrela, V.V. (2017). An Introduction
to Data Mining Applied to Health-Oriented Databases, Orient. J. Comp. Sci. and
Technol (OJCST), 9(3). DOI : 10.13005/ojcst/09.03.03
(15) Jesus,
M.A., Estrela, V.V., Saotome, O., & Stutz, D. (2018) Super-resolution via
particle swarm optimization variants. In: Hemanth J., Balas V. (Eds). Lecture
Notes in Computational Vision and Biomechanics, vol 25. Springer, Cham.
(16) Razmjooy, N., B. S. Mousavi,
Khalilpour, M. and Hosseini, H. “Automatic Selection and Fusion of Color Spaces
for Image Thresholding”. Signal,
Image and Video Processing, Vol. 8, no. 4 , 2014: 603-614.
(17) Razmjooy, N., Mousavi B.
S., Fazlollah S., Hosseini Khotbesara, M. “A Computer-Aided Diagnosis System
for Malignant Melanomas.” Neural
Computing and Applications, Vol. 23, no. 7-8, 2013: 2059-2071.
(18) Mousavi, B. S., and
Soleymani, F., Razmjooy, F. “Semantic Image Classification by Genetic Algorithm
Using Optimised Fuzzy System Based on Zernike Moments.” Signal, Image and Video Processing, Vol. 8, no. 5,
2014: 831-842.
(19) Hemanth, D.J., &
Estrela, V.V. (2017). Deep Learning for Image Processing Applications, Advances
in Parallel Computing Series, Vol. 31, IOS Press, ISBN 978-1-61499-821-1
(print), ISBN 978-1-61499-822-8 (online)