Feature extraction and segmentation of medical
images for MRI and Digital mammogram
Type of article: Original
Tinhinane Mehdi, Asma Boudrioua,
Abdelouhab Aloui
Medical
Computing Laboratory (LIMED), Faculty of Exact Sciences,University of Abderrahmane
Mira of Bejaia, Algeria.
Abstract:
Background: Developing a computer aided diagnosis
system (CAD) is an extremely challenging task. One of the major goals of CAD is
to help the radiologist to make good decisions by detecting and analyzing
characteristics of benign and malignant lesions. In this context, we present
accurate and automatic method that, detect and extract malignancy descriptors
of breast and meningioma brain tumor.
Methods:
We applied an algorithm that
uses enhancement image based on homomorphic filtering and adaptive histogram
equalization technique. A region of interest is determinate using K means clustering.
Then, we employed wavelet transform to extract pertinent features for
meningioma tumor, geometric and texture characteristics for breast tumor in
order to classify malignancy lesion.
Results: the segmentation result has been shown in
this paper showing the well segmented masses, as well as the extracted set of characteristics
has been illustrated in a vector.
Conclusion:
A features extraction and segmentation
of mass cancer and meningioma brain Tumor images are presented in this paper. Future work should focus on extraction of
pertinent information witch characterize the malignancy. In order to increase
the classification accuracy, we plan to explore a large data set of real
images. Then, to evaluate performance of our method, we will compare the recent
work proposed in literature.
Keywords: CAD; breast tumor; meningioma brain tumor; feature
extraction; segmentation.
Corresponding author: Ms Tinhinane Mehdi Faculty
of Exact Sciences, University of Abderrahmane Mira of
Bejaia, Algeria. Email: hinanemehdi20@gmail.com
Received: 15 July,
2018, Accepted: 02 January, 2019, English editing: 04 January, 2019, Published:
09 January, 2019.
Screened by iThenticate..©2017-2019 KNOWLEDGE KINGDOM PUBLISHING.
1. Introduction
Nowadays, cancer is a most eminent public health
problem in the world. According to the world health organization (WHO), breast
cancer and meningioma brain tumor are considered as most common primary cancer
in adults [1]. Moreover, they recur despite aggressive treatment, leading to
substantial morbidity. Standard-of-care management typically involves surgical
resection and often radiation therapy for high-grade.
Medical
image analysis plays an important role in assessing the spatial organization of
different tissues. Especially, the identification of tissue as normal, benign
or malignant. This classification can help clinicians by giving a second
opinion improving diagnostic accuracy and reduce human error.
Here, we present three steps of CAD system used for tumor
extraction for mammography images and MRI images. To achieve this, we propose a
method for pre-processing to delete noise and to improve the contrast between
different anatomical structures. Homomorphic filtering is applied to the input
image for improving the contrast of image. In addition, morphological
operations are applied to remove the noise and to smooth the edges of the
image. This method was inspired from work proposed in [2]. The K-Means
clustering algorithm and morphological operators has been used to segment mass
and extract the border witch is a both robust and successful approach. Finally,
we proceeded to the extraction of geometric and texture features for digital
mammograms and wavelet transform to extract pertinent features for meningioma
tumors.
2. Methods
The proposed segmentation model includes three main steps. The initial
step aims to delete noise and improve the contrast between different anatomical
structures. Homomorphic filtering, which is applied to the input image to
improve the contrast of image and morphological operations are applied to
remove the noise and to smooth the edges of the image. K-Means clustering
algorithm and morphological operators have been used to segment mass and
extract the border, which is robust and successful approach. Then, we proceeded
to the extraction of geometric and texture features for digital mammograms and
wavelet transform to extract pertinent features for meningioma tumor. In this
section, the different steps of the proposed method are explained. The
implementation of our model was performed on a data set of real patients.
Brain MRI data
The
dataset was acquired from the CHU La Cavale Blanche,
Brest-FRANCE and from haravad school site at https://nac.spl.harvard.edu/downloads.
The MRIs are sampled by 3.0 Tesla Philips Medical System. The dataset was scanned
by two different MRI modalities; the slices have dimensions of 256*256 Pixels.
Digital mammogram data
The mammogram used in the experiments is taken from
the mini mammography database of MIAS, where the masses are regrouped either
speculated, circumscribed or well-defined. The original MIAS Database (digitized
at 50 micron pixel edge) has been reduced to 200 micron pixel edge and
clipped/padded so that every image is 1024 pixels x 1024 pixels.
2.1 Preprocessing
Intensity
normalization is an important step in preprocessing of MR images. In this
Article we used an algorithm based on hybrid approach combination of both
frequency domain homomorphic filtering and a spatial domain morphology as
described by [3] and an adaptive histogram equalization technique to the output
of hybrid approach.
The steps of the approach are defined as follows:
step1: Apply
homomorphic filter to compress brightness range and enhance contrast of image.
Figure 1 shows the steps in homomorphic
filtering process

Fig.1.Homomorphic Filter
f(x,
y): an input image.
Z(x,
y) : output after log transformation. Z (u, v) is the output of Fourier
transform.
H (u, v) : transfer function of frequency domain filter.
H’ (u ,v) :output of the Z(u,
v) filtered with H(u,v). Exponential is applied to
the H’(x,y) to get the
output G (x, y).
H (u, v) = (rH - rL) [1-exp(c (D/D02)] Where rH is the
regulation parameter to change high frequency, rL is
regulation of parameter to change low frequency, where rH>1
and rL<1, c is sharpening parameter and D is
balance parameter. rH =1.414 and rL=0.18.
D (u, v) =u2+v2, D0 is harmonic coefficient, D0 = ((max-µ)2+ (min-µ)2)/ a
Step2: Tophat
transform is applied to G using disk of radius 15 as a structuring element.
Shape and size of structuring element is selected based on the shape and size
of the masses. It can be used to separate the objects. Let the output be thf.
Step 3: Dilation operator on a binary image is
to gradually enlarge the boundaries of regions of foreground pixels. It is applied to smooth the borders of tophat transformed image. Let the output be thf1.
Step4: Bothat
transform is applied to the original image to smooth the objects in original
image. Let the output be bhf.
Step 5: These images are combined
using Image arithmetic addition and subtraction.
Enhanced image = (G+thf1) – (bhf)
Step6: Adaptive histogram equalization
technique is applied to improve local contrast.
2.2 Segmentation
To detect
automatically the tumor and treat only the region of interest and not all pixels
in images we used the K-Means clustering algorithm and morphological operators
to segment mass and extract the edge.
The k means-clustering algorithm uses iterative refinement to produce a
final result. The algorithm inputs are the number of clusters and the data set
input is image pixels and their descriptor are their grey level values.
Euclidean distance has been chosen as distance.
2.3 Feature Extraction
For brain tumor, the wavelet transform has become a major image
characterization tool. We used Discrete Wavelet Transform to extract pertinent
features that describe malignant lesion. The reduced features were submitted.
In fact, according to the BIRADS, benign masses have a round or oval
shape and a circumscribed or micro lobulated contour, while malignant masses
usually have a lobed or irregular shape and an indistinct or speculated
contour. Therefore, we extract geometric features such as: Circularity Index,
Area, Perimeter P: A: is the ratio of
perimeter to the area of the lesion. L: S Ratio: is the length ratio of the
major (long) axis to the minor (short) axis of the equivalent ellipse of the
lesion. If L: S ratio is more, it is likely the lesion is malignant. E: N
(Elliptical normalized circumference): Anfractuosity is a common morphological
feature for malignant contour. ENC is the circumference ratio of the lesion and
its equivalent ellipse. Anfractuosity of a lesion contour is characterized by
ENC. and we opted for the extraction of texture descriptor that are important
for the distinction of the normal tissue from the cancerous one, for this we
use a gray level co-occurrence matrix.
The steps of the approach are defined in Fig2.
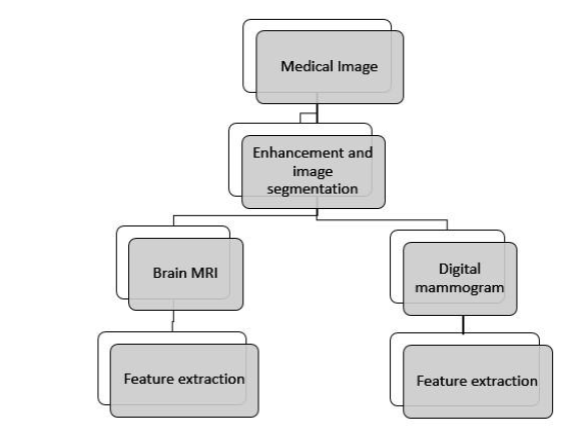
Fig.2. Proposed method
3. Results and discussion
In this section, we present the results of our
approach, starting with the preprocessing which is illustrated in Fig 3.
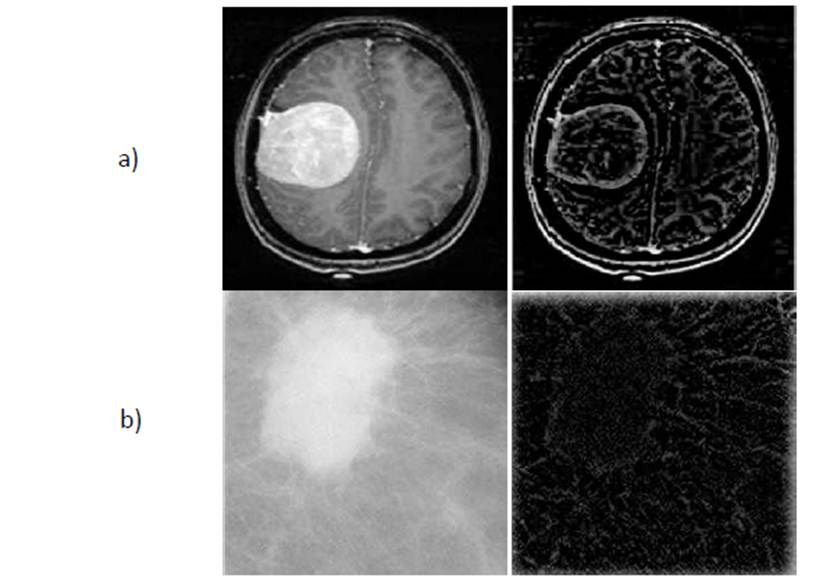
Fig 3. a) Original
and Enhaced MRI b) Original Mammogram and Enhaced Mammogram.
Then the ROI’s determination are given in the
figure below,
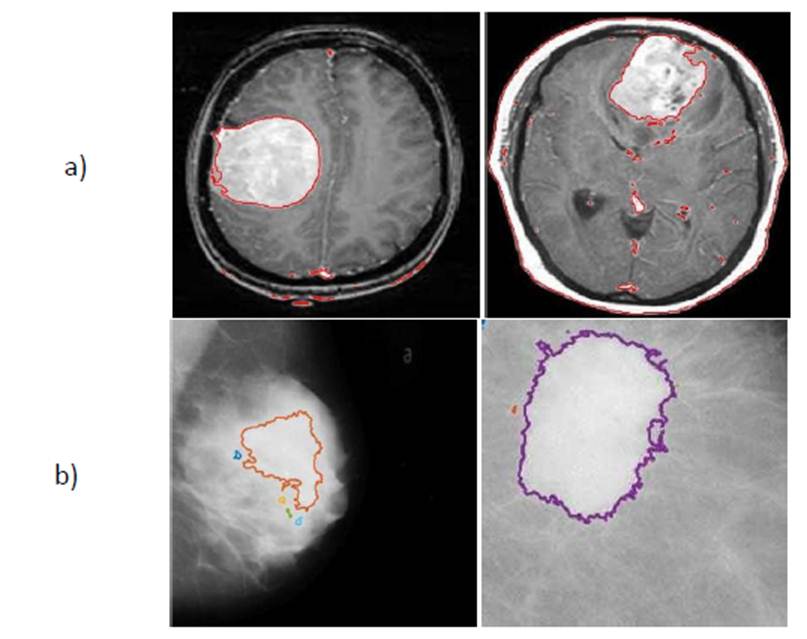
Fig.4.a) Result of brain lesion b) Result of breast lesion.
The following
table shows the values of the extracted descriptors.
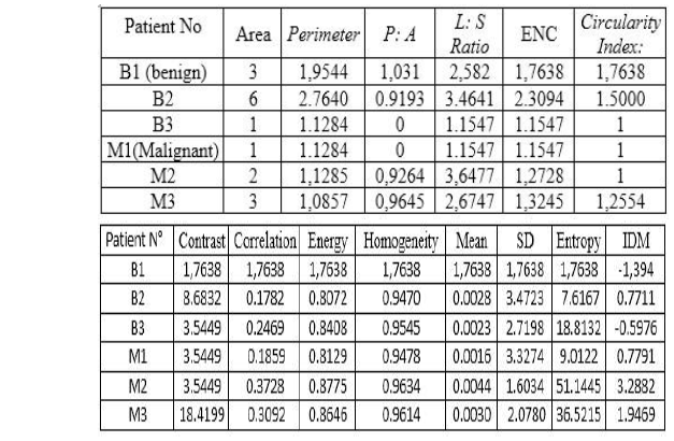
Fig 5. Vector characterization
The purpose of our work is to have good preprocessing results
and efficient descriptors in order to obtain better classification model, we
aim to compare our results through many classifiers like KNN and SVM
essentially with the machine learning.
4. Conclusion
A features extraction and segmentation of mass cancer
and meningioma brain Tumor images are presented in this paper. The proposed
method is mainly based on Zhang et al. [3] Future work should focus on
extraction of pertinent information that characterize the malignancy. In order
to increase the classification accuracy, we plan to explore a large data set of
real images. Compare our results with machine learning classifiers to find the
most appropriate one for our approach.
5. Conflict of interest statement
We certify that there
is no conflict of interest with any financial organization in the subject
matter or materials discussed in this manuscript.
6. Authors’ biography
No Biography
7. References
[1]Arianna Mencattini et al. Automatic breast masses
boundary extraction in digital mammography using spatial fuzzy c-means
clustering and active contour models, IEEE International Workshop on Medical
Measurements and Applications Proceedings (MeMeA),
2011, pp.632637.
[2]Spandana Paramkusham, K. M. M. Rao, B. V. V. S. N. Prabhakar Rao, Automated Approach for Qualitative
Assessment of Breast Density and Lesion Feature Extraction for Early Detection
of Breast Cancer, TECHNIA, International Journal of Computing Science and
Communication Technologies, vol.6, No.1, July. 2013 (ISSN 0974- 3375)
[3]Zhang Chaofu, MA Li-ni, Jing Lu-na, Mixed Frequency domain and spatial of enhancement algorithm
for infrared image, IEEE international Conference on Fuzzy Systems and
Knowledge Discovery, FSKD, 2012, pp:2706-2710.
[4]Li et al.Breast masses in mammography classifcation with local contour features BioMedical Engineering OnLine DOI
10.1186/s12938-017-0332-0,2017, 16:44.
[5] JSaeed Khodary et all. Enhancement Accuracy of Breast Tumor
Diagnosis in Digital Mammograms, Journal of Biomedical sciences; vol.6, No.4:28,
August 17, 2017(ISSN 2254-609X).
[6]Anamika Ahirwar, Measure the Effectiveness of an
Innovative Scheme for Medical Imaging, International Journal of Computer
Applications (0975 8887) Volume 37 No.2, January 2012.
[7]Ramani, R. G., & Sivaselvi, K. Classification of
Pathological Magnetic Resonance Images of Brain Using Data Mining Techniques.
In Recent Trends and Challenges in Computational Models (ICRTCCM), 2017 Second
International Conference on (pp. 77-82). IEEE. February 2017,
[8]Coroller, T. P., Bi, W. L., Huynh, E., Abedalthagafi,
M., Aizer, A. A., Greenwald, N. F.,, &Gupta, S.
(2017), Radiographic prediction of meningioma grade by semantic and radiomic features. PloS one,
12(11), e0187908. https://doi.org/10.1371/journal.pone.0187908
[9] spanhol, Fabio A.,oliveira , Luiz S., petitjeanPETITJEAN, Caroline, et al. A dataset for breastcancer histopathological image classification. IEEE
Transactions on Biomedical Engineering, 2016, vol. 63, no 7, p. 1455-1462