Semantic Segmentation of
Medical Images with Deep Learning: Overview
Type of article: Review
Yamina Azzi1, Abdelouahab
Moussaoui1, Mohand- Tahar Kechadi 2
1 Department of Computer science at Faculty of science
Ferhat Abbas University Sétif, Algeria.
2 School of Computer Science, University
College Dublin, Ireland.
Abstract
Semantic
segmentation is one of the biggest challenging tasks in computer vision, especially in medical image analysis, it helps to locate
and identify pathological structures automatically. It is an active research
area. Continuously different techniques are proposed. Recently Deep Learning is
the latest technique used intensively to improve the performance in medical
image segmentation. For this reason, we present in this non-systematic review a
preliminary description about semantic segmentation with deep learning and the
most important steps to build a model that deal with this problem.
Keywords: Semantic
segmentation, Deep Learning, Medical images, Segmentation.
Corresponding author: Yamina Azzi Department of Computer science at
Faculty of science Ferhat Abbas university Sétif, Algeria
Email: yamina.azzi@univ-setif.dz
Received: July 12 2020.
Reviewed: August 17 2020. Accepted: October 3 2020. Published: December 7
2020.
Medical Technologies
Journal subscribes to the principles of the Committee on Publication Ethics
(COPE).
Screened by iThenticate..©2017-2020 KNOWLEDGE KINGDOM PUBLISHING.
1.
Introduction
In the
healthcare sector, medical imaging is an essential protocol for many disease
diagnostics, treatment planning, and patients monitoring, however medical images
suffer from many problems related to their weak resolution and consequently the
interpretation may be a tedious task and time consuming for human experts. Automatic
segmentation of medical images requires high precision that is why it is a very
challenging task due to the large variation in the anatomy shapes of patients
and the low contrast between tissues [1].
Traditional
image segmentation techniques are based on identifying the objects in the image
by detecting contours or locating regions using a variety of rigid algorithms.
Modern image segmentation techniques are powered by Machine Learning ML, which is a branch of Artificial Intelligence AI. This last means the
ability to learn from data and improve without being programmed. In the context
of image segmentation, it consists to define a mathematical model that learns
how to segment a set of images known as training set. This technique extracts
the useful features for enabling the segmentation of new unseen images. The
learning process is ether supervised or unsupervised [1-5].
With the
Supervised learning, the training images are associated with labels that
represent the true segmentation of training images. These labels are provided
by human experts. The machine-learning algorithm builds the model progressively
by rectifying the weights of features to obtain the expected output values.
However,
in the case of Unsupervised Learning, the training images are presented without
labels. The model should segment images and learn structures from the training
data set automatically, without any human guidance. The human expert
interpretation is requisite only at the end.
The new term of
semantic segmentation is related to the last trending technique in machine learning:
Deep Learning. It consists of using a supervised learning and a labeled
dataset. Recently it is the most frequently used technique in medical image
segmentation. It provides very exciting results, offering better performance
and high precision in locating pathological structures [2-].
In the next sections,
we are going to present the definition of image segmentation with some examples
of techniques used before deep learning. Then we describe the new aspect of
semantic segmentation with more details about the Deep Learning technique. The
aim of this paper is to review the general steps of semantic segmentation with
Deep Learning. We highlighted the most frequently implemented architectures in
this context, after that we expose some deep learning application results in
brain tumor segmentation on the Brats dataset.
2.
Discussion
2.1 Image segmentation
Image
segmentation is the process of dividing an image into regions or objects that
are located in, to understand it in a much-grained level. It helps to identify
and locate pathological structures in the case of medical images such as
tumors, fractures and bruises bones or blood hemorrhages.
The following three types are the most notable
techniques used for this purpose:
i.
Contour based segmentation or edge detection:
are the primary and the naive techniques used in this area where the aim is to
identify boundaries in the image by applying filters that detect the intensity
variation or discontinuities created by the contour’s pixels, the frequently
used filters are sobel, prewitt, laplacian and canny detectors [1].
ii.
Region based segmentation: encompasses
a set of techniques that aim to identify areas of different objects in the same
image based on the similarity criterion between pixels within each region,
there are various approaches such as split and merge algorithm and region
growing algorithm
iii.
Segmentation based on clustering: is unsupervised machine learning technique, where
the goal is to partition an image into a set of finite categories known as
clusters by classifying pixels automatically without knowing classes in
advance, a similarity measure like Jaccard coefficient or Euclidean distance is
defined between pixels, then similar pixels are grouped together to form the
set of clusters. The grouping of pixels is based on the principle of maximizing
the intra-class similarity and minimizing the inter-class similarity. There are
several clustering algorithms as hard clustering; k- means clustering, fuzzy
clustering
2.2
Semantic segmentation with deep learning
Semantic
segmentation is the same concept of the ordinary image segmentation the only
difference is that pixels are classified semantically to an appropriate class
to form regions, it offeres a better
understanding of the image context [14]. Deep Learning is a machine
learning technique bio-inspired from human brain. It is in the form of a neural
network where simple neural network consists of input and output layers
interconnected by just one hidden layer, but in Deep Neural Network multiple
hidden layers interconnect them [2-3].
In the context of semantic segmentation with
deep learning, the learning process is supervised where the training images are
associated with the labels, which are the training images with true-segmented
classes called also ground truth data.
The deep
learning algorithm takes the training images as an input. It consists of applying
a set of operations that help to extract features. After that, it consists of performing
a predicted segmentation, which will be compared with the ground truth. An error
function is applied to estimate the efficiency. A back propagation algorithm is
applied to tune the network weights and minimize the error. The algorithm
trends to obtain the convergence towards the best accuracy and segmentation
precision [16-17].
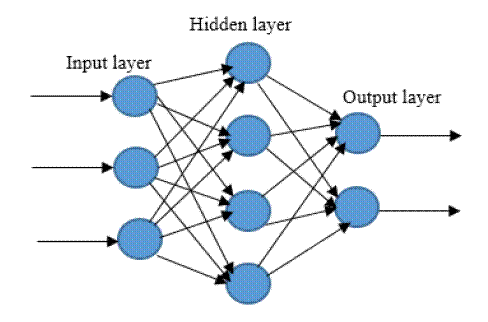
Figure.1 Simple Neural Network
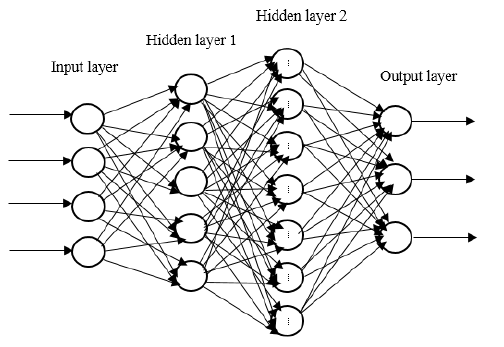
Figure.2 Deep Neural Network
2.3 Concepts
These two
concepts are the basic recommended algorithms to start building an image
segmentation model using deep learning [18-20].
·
Convolutional neural network (CNN): is a
deep neural network algorithm used to deal with the image classification
problems. It consists of two main paths: feature extraction path and
classification path. The feature extraction path, also known as a down-sampling path, is made of a series of
convolutional and pooling layers during which useful features are extracted. Convolution
layer uses a set of learnable filters applied over the input images to build
the feature map. The pooling layer used to reduce the spatial dimension of the
feature map to get more information about what happen in the image and to gain
better computation performance. In the classification task, the down-sampling
path must be connected to a fully connected layer that takes the output feature
map to generate the probabilities for the object in the image to predict the
appropriate label [4, and 14].
·
Fully convolutional neural network (FCN): is similar
to convolution neural network
architecture, but instead of the fully connected layer in the classification
path. FCN uses an up-sampling
path, which consisted of layers with opposite operations that are in the
down-sampling path. These operations may be the reverse operations of the
convolution (transposed convolution) or the pooling operations (unpooling with nearest
neighbor interpolation or bed of nails). In addition to some skip connection
which is a kind of merging and concatenating information between layers of the two
paths in order to recover the spatial information of the objects ”where are
this objects located”. At the end, an output image with a set of probabilities
is generated to identify the classes [5-6].
Down-sampling
in the feature extraction path causes significant loss in the spatial dimension
of objects and in the CNN the fully connected layers doesn’t allow the recovery
of this loss, but in the FCN the up-sampling path compensates this loss and helps to reconstruct the input
image with the objects segmentation. That is why this architecture is widely used
in semantic segmentation task.
2.4 Input
The input of
the model represents a
set of regular grayscale medical images in 2D or 3D, sometimes they may suffer
from noise because of some artifacts during acquisition phase. So before being
diagnosed they go through some processing techniques that ensure the quality
like denoising using median filter. For deep learning technique, the uniformity
of the training data is a major task that quickens the learning process and
avoids the domination of features on large scale.
The following
techniques are frequently used to achieve better performance:
– Min-max scale: is
a normalization technique aiming to scale data into the range [0, 1] by
computing the maximum and the minimum of the data, and then subtracting the
minimum from the data and dividing the results by the obtained result of
subtraction between the maximum and the minimum.
– Z-score standardization: is
a strategy of normalizing data by subtracting the mean and dividing by the standard
deviation.
– Histogram of equalization: is
a technique for adjusting image pixels intensities and perform contrast
enhancement [7-9].
2.5 Ground truth images
In this context,
ground truth image (image label) represents an image with the same size and the
same information to the corresponding image in the training set where each
pixel intensity is associated to the appropriate category or class label. The
model compares the predicted output segmentation with the label image in order
to adjust the weights and minimize the error between them to obtain a precise localization
for each object with the exact semantics. For medical grayscale images used in
the training process the labels can be presented in one of these two forms:
class label or one-hot-vector-encoding
1. Class label format:
each class is labeled by a definite integer number so each pixel in the ground
truth will take the value of a class as intensity value.
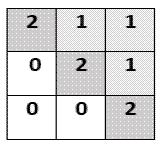
Fig. 3
Class label format
2. One-hot-vector
encoding: is a
representation of categorical class labels as binary vectors. It requires that
the class category be integer. Then each class will be converted to a binary
vector of size (height*width of the original image), where all values are zero
except the index of the class will take 1. This representation is the most
compatible with deep learning frameworks and in general, the class label format
is converted implicitly or explicitly to one hot encoding before processing.
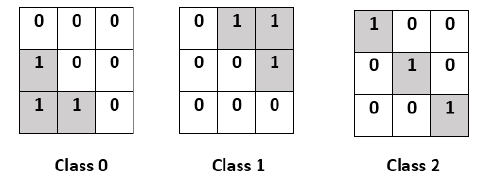
Fig. 4 One hot-vector encoding
2.6 Output
The output of
the deep learning model for semantic segmentation is a set of images equal to the training set images,
with depth equivalent to the number of classes, where each channel represents label
probabilities in the image. For binary classes a sigmoid function is used to
generate the class probabilities, but in the case of multiclass segmentation, a
softbacks function is used to
generate the probabilities for each pixel. Finally, a similarity measure is used in order to measure how the
output and label images are similar, and compute the loss between them. Then
the back propagation algorithm is
used to adjust the weights that maximize the similarity and minimize the loss to improve the segmentation performance.
For example, one of the most powerful similarity measures applied in medical image segmentation is the Dice score
coefficient.
2.7 Medical images application examples and results
Gliomas are the most aggressive primary tumors that develop in the human
brain and can menace human life. It can appear into two grades: 1) low-grade
gliomas are most of time curable and characterized by high survival rate up to
10 years or high-grade gliomas, which are incurable, and 2) the proposed treatment
can just reduce pain and improve survival rate to 2-5 years. In order to decide
the type of gliomas and the treatment plan the tumor part should be well
identified and this task is very hard by handcrafting so automatic segmentation
with deep learning is widely used in these tasks, and is proving its
effectiveness in term of precision.
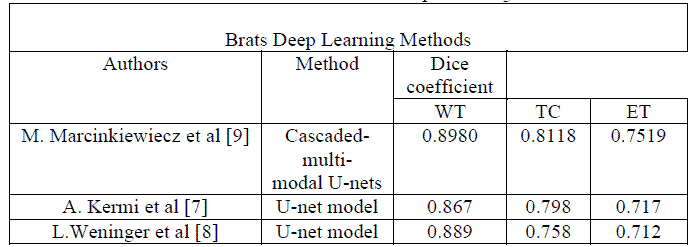
In the table 1,
we summary some results of deep learning segmentation models tested on the
Brats 2018 validation set. The models were trained on Brats 2018 training set, which
contains 210 patients with high-grade gliomas (HGG/Glioblastoma) and 75 with low-grade
gliomas (LGG). Each patient has 4 MRI scans with different modalities
T1weighted, T2 weighted, Flair(Fluid attenuated inversion recovery), T1ce (T1 contrast-enhanced)
with the ground-truth for gliomas segments, the gliomas structures are rated
with three integer rates 1 for necrotic/non enhancing tumor, 2 for edema, 4 for
enhancing tumor and 0 for everything else [10-11]. All the proposed methods are
based on a customized FCN architecture, which was efficient in biomedical image
segmentation called U-net [12-14].
Table1:
Published results of Brats
Deep Learning Methods
Where:
– WT: whole tumor includes all tumor structures
(1+2+4 labels).
– TC: tumor core includes all tumor structures
except edema (1+4).
– ET: enhancing tumor (label 4) as figure 5 shows.
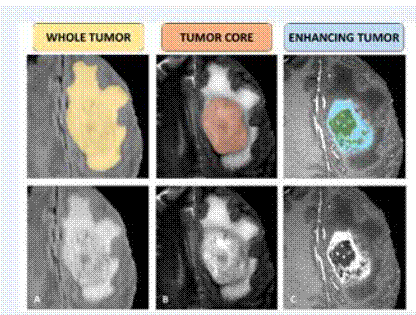
Fig.5. Brain tumor
parts Brats dataset.
3.
Conclusion
In
this survey, we presented an overview about image segmentation and the past
techniques used for this purpose, until the semantic segmentation with deep
learning in the medical imaging field. We introduced the general structure to
follow in build the model. We exposed the most effectiveness architectures such
as convolutional neural networks, fully convolutional neural network, and u-net.
, we presented some important information about input and output data for the
models. Finally, we provided some results of accurate model in MRI brain tumors
semantic segmentation using deep learning for high-grade gliomas and low-grade
gliomas.
4.
Conflict of interest statement
We
certify that there is no conflict of interest with any financial organization
in the subject matter or materials discussed in this manuscript.
5.
Authors’ biography
Yamina
Azzi
PhD student at Department of Computer science at Faculty of
science Ferhat Abbas University Sétif, Algeria.
Abdelouhab Moussaoui
Professor at Department of
Computer science at Faculty of science Ferhat Abbas University Sétif, Algeria.
Mohand Tahar Kechadi
Professor at School of Computer Science, University College Dublin,
Ireland.
6.
References
[1] Song yuheng , yan hao, «Image segmentation algorithms
overview,» Asia Modelling Symposium (AMS),IEEE, p. 6, 2017. https://doi.org/10.1109/AMS.2017.24
[2] Lee, JuneGoo; Sanghoom Jun,
Young-Won Cho,Hyunna Lee,Guk Bae Kim, Joon Beom Seo,NamKug Kim, «Deep learning
in medical imaging : general overview,» Koreanjournal of radiology,
2017.https://doi.org/10.3348/kjr.2017.18.4.570 PMid:28670152 PMCid:PMC5447633
[3] Geert Litjens, Thijs Kooi, Babak
Ehteshami Bejnordi, Arnaud Arindra Adiyoso Setio, Francesco Ciompi,, «A Survey
on Deep Learning in Medical Image Analysis,» Medical Image Analysis, vol. vol,
n° %142, pp. 60-88, Décembre 2017. https://doi.org/10.1016/j.media.2017.07.005 PMid:28778026
[4] Baris Kayalibay, Grady Jensen, Patrick van
der Smagt, «CNN-based Segmentation of Medical Imaging Data,» arXiv:1701.03056v2
[cs.CV], 25 Jul 2017.
[5] Zhao X, Wu Y, Song G, Li Z, Zhang Y, Fan
Y,, «A deep learning model integrating FCNNs and CRFs for brain tumor
segmentation,» Medical Image Analysis, vol. vol, n° %143, pp. 98-111, January
2018. https://doi.org/10.1016/j.media.2017.10.002 PMid:29040911 PMCid:PMC6029627
[6] Patrick Ferdinand Christ, Florian
Ettlinger, Felix Grün, Mohamed Ezzeldin A. Elshaera, Jana Lipkova, Sebastian
Schlecht, Freba Ahmaddy, Sunil Tatavarty, Marc Bickel, Patrick Bilic, Markus
Rempfler, Felix Hofmann, Melvin D Anastasi, Seyed-Ahmad Ahmadi, Geo, «Automatic
Liver and Tumor Segmentation of CT and MRI Volumes Using Cascaded Fully
Convolutional Neural Networks,» arXiv:1702.05970v2, 2017.
[7] Kermi A., Mahmoudi I., Khadir M.T., «Deep
Convolutional Neural Networks Using U-Net for Automatic Brain Tumor
Segmentation in Multimodal MRI Volumes,» In: Crimi A., Bakas S., Kuijf H.,
Keyvan F., Reyes M., van Walsum T. (eds) Brainlesion: Glioma, Multiple
Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2018. Lecture Notes in
Computer Science, vol. vol, n° %111384 Springer, Cham, 2019.
https://doi.org/10.1007/978-3-030-11726-9_4
[8] Weninger L., Rippel O., Koppers S., Merhof
D., «Segmentation of Brain Tumors and Patient Survival Prediction: Methods for
the BraTS 2018 Challenge,» In: Crimi A., Bakas S., Kuijf H., Keyvan F., Reyes
M., van Walsum T. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and
Traumatic Brain Injuries. BrainLes 2018. Lecture Notes in Computer Science,
Vols. %1 sur %2vol 11384. Springer, Cham, 2019.
https://doi.org/10.1007/978-3-030-11726-9_1
[9] Marcinkiewicz M., Nalepa J., Lorenzo P.R.,
Dudzik W., Mrukwa G,, «Segmenting Brain Tumors from MRI Using Cascaded
Multi-modal U-Nets,» In: Crimi A., Bakas S., Kuijf H., Keyvan F., Reyes M., van
Walsum T. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic
Brain Injuries. BrainLes 2018. Lecture Notes in C, Vols. %1 sur %2vol 11384.
Springer, Cham. https://doi.org/10.1007/978-3-030-11726-9_2
[10] S. Bakas, M. Reyes, A. Jakab,
S. Bauer, M. Rempfler, A. Crimi, et al.,, «"Identifying the Best Machine
Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and
Overall Survival Prediction in the BRATS Challenge",,» arXiv preprint
arXiv:1.
[11] B. H. Menze, A. Jakab, S.
Bauer, J. Kalpathy-Cramer, K. Farahani, J. Kirby, et al., «"The Multimodal
Brain Tumor Image Segmentation Benchmark (BRATS)",» , IEEE Transactions on
Medical Imaging 34(10), 1993-2024 (2015) DOI: 10.1109/TMI.2014.2377694. https://doi.org/10.1109/TMI.2014.2377694 PMid:25494501 PMCid:PMC4833122
[12] Ronneberger, O.; Fischer, P.;
and T. Brox, «U-net: Convolutional networks for biomedical image segmentation,»
arXiv:1505.04597v1 [cs.CV], 2015. https://doi.org/10.1007/978-3-319-24574-4_28
[13] Zhang, Hejia; Zhu, Xia; Willke,
Theodore L., «Segmenting Brain Tumors with Symmetry,»
arXiv:1711.06636v1[cs.CV], 2017.
[14] Saddam Hussain, Syed Muhammad
Anwar, Muhammad Majid,, «Segmentation of Glioma Tumors in Brain Using Deep
Convolutional Neural,» arXiv, 1 Aug 2017. https://doi.org/10.1016/j.neucom.2017.12.032
[15] Salma Alqazzaz, Xianfang Sun,
Xin Yang, Len Nokes1, «Automated brain tumor segmentation on multi-modal MR
image using SegNet».
[16] Ali Isin a, Cem
Direkoglu,Melike sah,, «Review of mri-based brain tumor image segmentation
using deep learning methods,,» Elsevier, , 30 august 2016. https://doi.org/10.1016/j.procs.2016.09.407
[17] Sanghoom Jun,Young-Won
Cho,Hyunna Lee,Guk Bae Kim,Joon Beom Seo,NamKug, «Deep learning in medical
imaging : general overview.,» Koreanjournal, 18/04/2017. https://doi.org/10.3348/kjr.2017.18.4.570 PMid:28670152 PMCid:PMC5447633
[18] van der Laak,Bram van Ginneken,Clara
I,Sanchez Mohsen ghafoorian,Jeroen, «A survey on deep learning in medical image
analysis.,» Elsevier, 2017.
[19] A. Garcia-Garcia, S. Orts-Escolano, S.O.
Oprea, V. Villena-Martinez, and J. Garcia-Rodriguez, «A Review on Deep Learning
Techniques Applied to Semantic Segmentation,» arXiv:1704.06857v1 [cs.CV] , 22
Apr 2017.
[20] Holger R. ROTH,Chen SHEN, Hirohisa
ODA,Masahiro ODA,Yuichiro HAYASHI,Kazunari MISAWA,Kensaku MORI, «Deep learning
and its application to medical image segmentation,» arXiv:1803.08691v1 [cs.CV]
, 23 Mar 2018.